Dataset :
It is given by Kaggle from UCI Machine Learning Repository, in one of its challenges. It is a dataset of Breast Cancer patients with Malignant and Benign tumor.
K-nearest neighbour algorithm is used to predict whether is patient is having cancer (Malignant tumour) or not (Benign tumour).
Implementation of KNN algorithm for classification.
Code : Importing Libraries
# performing linear algebra import numpy as np # data processing import pandas as pd # visualisation import matplotlib.pyplot as plt |
Code : Loading dataset
df = pd.read_csv("..\breast-cancer-wisconsin-data\data.csv") print (data.head) |
Output :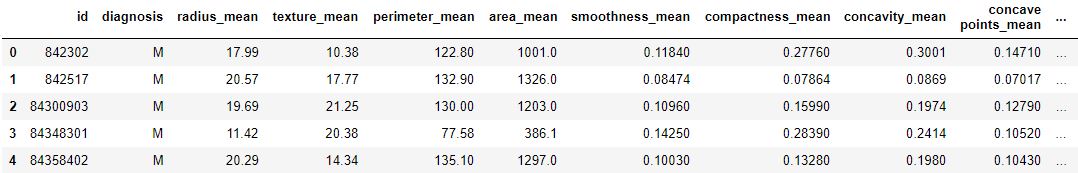
Code: Data Info
df.info() |
Output :
RangeIndex: 569 entries, 0 to 568 Data columns (total 33 columns): id 569 non-null int64 diagnosis 569 non-null object radius_mean 569 non-null float64 texture_mean 569 non-null float64 perimeter_mean 569 non-null float64 area_mean 569 non-null float64 smoothness_mean 569 non-null float64 compactness_mean 569 non-null float64 concavity_mean 569 non-null float64 concave points_mean 569 non-null float64 symmetry_mean 569 non-null float64 fractal_dimension_mean 569 non-null float64 radius_se 569 non-null float64 texture_se 569 non-null float64 perimeter_se 569 non-null float64 area_se 569 non-null float64 smoothness_se 569 non-null float64 compactness_se 569 non-null float64 concavity_se 569 non-null float64 concave points_se 569 non-null float64 symmetry_se 569 non-null float64 fractal_dimension_se 569 non-null float64 radius_worst 569 non-null float64 texture_worst 569 non-null float64 perimeter_worst 569 non-null float64 area_worst 569 non-null float64 smoothness_worst 569 non-null float64 compactness_worst 569 non-null float64 concavity_worst 569 non-null float64 concave points_worst 569 non-null float64 symmetry_worst 569 non-null float64 fractal_dimension_worst 569 non-null float64 Unnamed: 32 0 non-null float64 dtypes: float64(31), int64(1), object(1) memory usage: 146.8+ KB
Code: We are dropping columns – ‘id’ and ‘Unnamed: 32’ as they have no role in prediction
df.drop(['Unnamed: 32', 'id'], axis = 1) print(df.shape) |
Output:
(569, 31)
Code: Converting the diagnosis value of M and B to a numerical value where M (Malignant) = 1 and B (Benign) = 0
def diagnosis_value(diagnosis): if diagnosis == 'M': return 1 else: return 0 df['diagnosis'] = df['diagnosis'].apply(diagnosis_value) |
Code :
sns.lmplot(x = 'radius_mean', y = 'texture_mean', hue = 'diagnosis', data = df) |
Output:
Code :
sns.lmplot(x ='smoothness_mean', y = 'compactness_mean', data = df, hue = 'diagnosis') |
Output: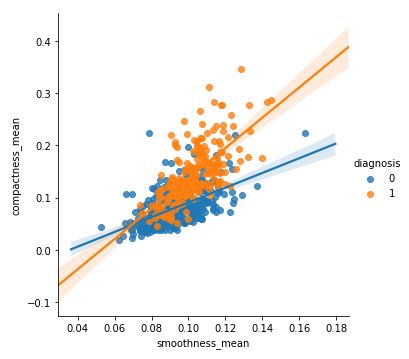
Code : Input and Output data
X = np.array(df.iloc[:, 1:]) y = np.array(df['diagnosis']) |
Code : Splitting data to training and testing
from sklearn.model_selection import train_test_split X_train, X_test, y_train, y_test = train_test_split( X, y, test_size = 0.33, random_state = 42) |
Code : Using Sklearn
knn = KNeighborsClassifier(n_neighbors = 13) knn.fit(X_train, y_train) |
Output:
KNeighborsClassifier(algorithm='auto', leaf_size=30,
metric='minkowski', metric_params=None,
n_jobs=None, n_neighbors=13, p=2,
weights='uniform')
Code : Prediction Score
knn.score(X_test, y_test) |
Output:
0.9627659574468085
Code : Performing Cross Validation
neighbors = [] cv_scores = [] from sklearn.model_selection import cross_val_score # perform 10 fold cross validation for k in range(1, 51, 2): neighbors.append(k) knn = KNeighborsClassifier(n_neighbors = k) scores = cross_val_score( knn, X_train, y_train, cv = 10, scoring = 'accuracy') cv_scores.append(scores.mean()) |
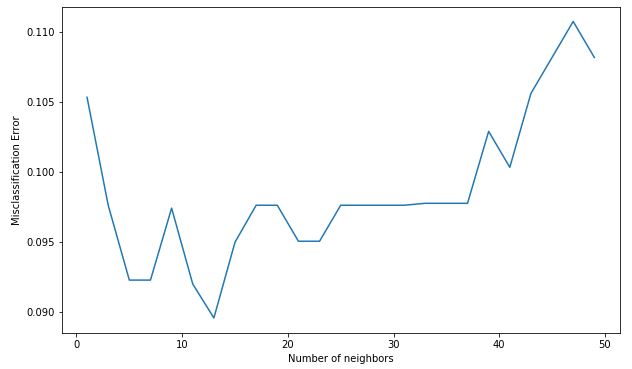
Code : Misclassification error versus k
MSE = [1-x for x in cv_scores] # determining the best k optimal_k = neighbors[MSE.index(min(MSE))] print('The optimal number of neighbors is % d ' % optimal_k) # plot misclassification error versus k plt.figure(figsize = (10, 6)) plt.plot(neighbors, MSE) plt.xlabel('Number of neighbors') plt.ylabel('Misclassification Error') plt.show() |
Output:
The optimal number of neighbors is 13